W-ASAP: Building Switzerland's Viral Wastewater Surveillance Platform
W-ASAP: Building Switzerland's Viral Wastewater Surveillance Platform
Infrastructure for National-Scale Genomic Analysis
Working Period: October 2024 - January 2026
On January 30, 2026, I left ETH Zurich's D-BSSE after 17 months as Software Engineer building W-ASAP (Wastewater Analysis & Sharing Platform) - the first of its kind read-level online tool for wastewater early viral detection.
Wastewater As Soon As Possible
Delivering on the promise of wastewater surveillance: early variant detection at national scale - the only financially sustainable path to population-wide viral monitoring.
The Challenge
Wastewater surveillance is a powerful tool for early detection of viral variants circulating in a population. Switzerland, like many countries, sequences viral fragments from wastewater treatment plants nationwide. But turning raw sequencing data into actionable insights for public health teams was a bottleneck.
The status quo:
- Cluster-only access: Analysis required running complex workflows on ETH's Euler HPC cluster, accessible only to bioinformatics experts
- Days of turnaround: Researchers had to request data analysis from a small group with cluster access and internal knowledge
- Isolated data: Wastewater data couldn't be directly compared with clinical variant definitions without manual, time-consuming steps
The goal was to transform this into something accessible to virologists, epidemiologists, and public health teams - without requiring terminal expertise.
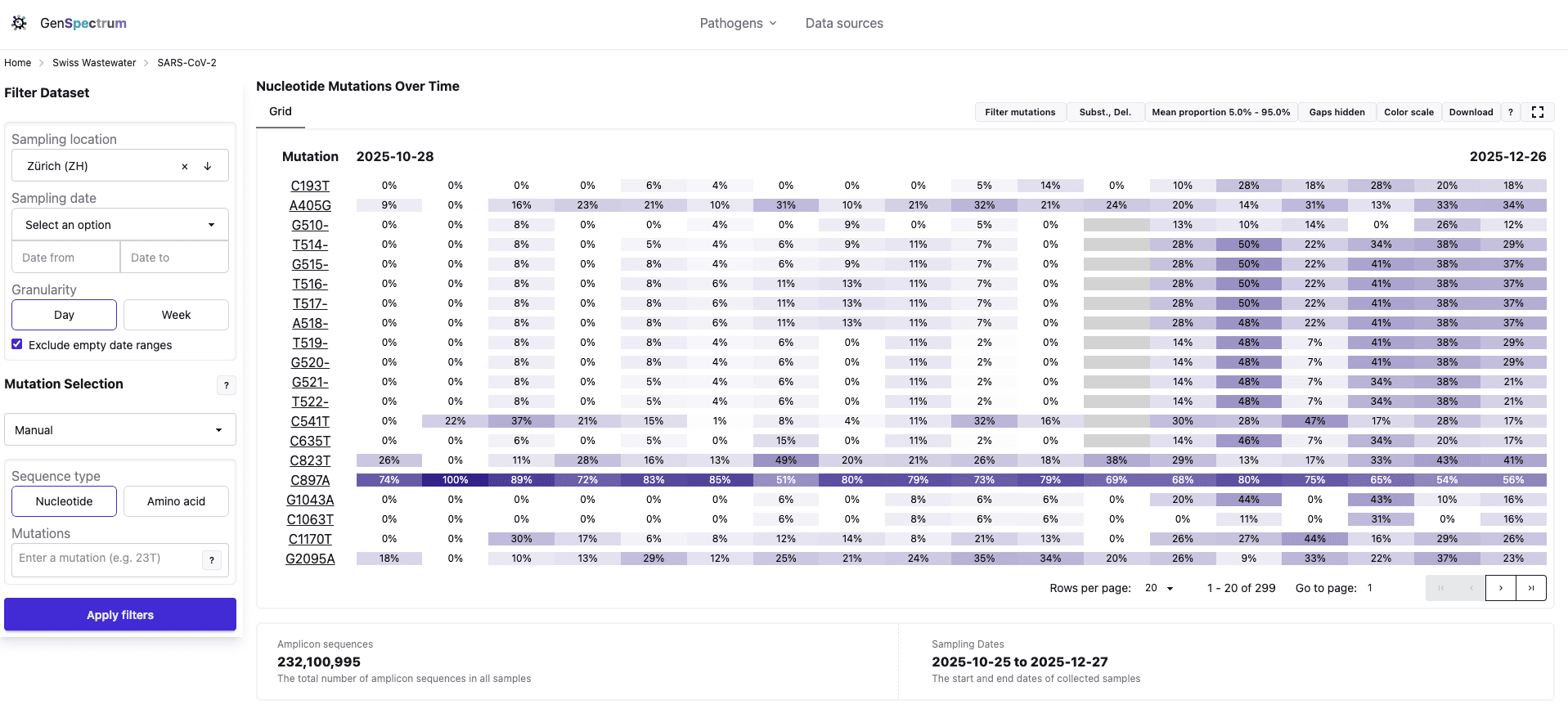
The Solution: Browser-Based Variant Exploration
W-ASAP bridges the gap between cluster-based terminal workflows and browser-based interactive exploration. It leverages existing infrastructure while making the data accessible:
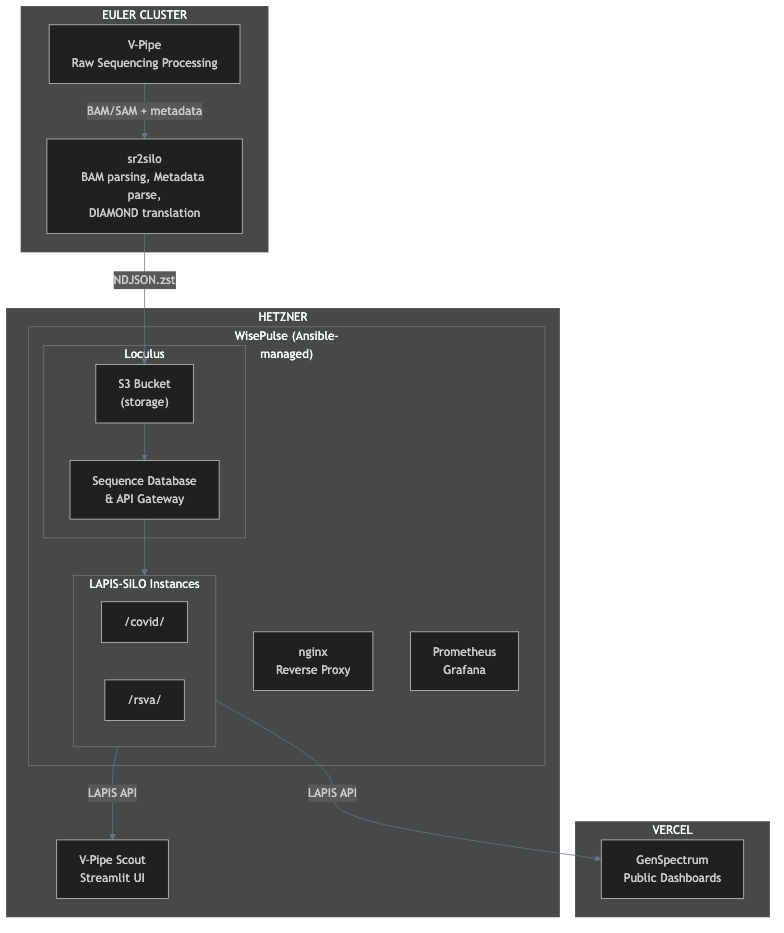
Data Flow
- V-Pipe (bioinformatics pipeline) processes raw sequencing data on ETH's Euler cluster
- sr2silo converts genomic alignment files for query engine ingestion
- Data submitted to Loculus (sequence database) via S3 bucket
- WisePulse (Ansible) ingests data into per-virus LAPIS-SILO (genomic query engine) databases
- LAPIS-SILO instances serve queries via REST API
- GenSpectrum (pathogen analysis platform) dashboards provide public web interfaces
The key innovation: first application of LAPIS-SILO to short read sequencing data. LAPIS-SILO, built by the GenSpectrum team, is a high-performance genomic query engine previously used for consensus-level data. Applying it to read-level wastewater data required careful engineering of the data pipeline and memory optimization.
What I Built
I took this project from concept to proof-of-concept to production. Three main components:
sr2silo - Python ETL pipeline converting genomic alignments for database ingestion. Published on Bioconda, runs daily on ETH's Euler cluster.
WisePulse - Ansible-based infrastructure as code for all W-ASAP services. Multi-virus pipeline with automatic rollback. Adding new viruses is now configuration, not engineering.
V-Pipe Scout (live demo) - Streamlit prototyping frontend for variant exploration. Served as testing ground before GenSpectrum integration.
Core Technology: LAPIS-SILO
github.com/GenSpectrum/LAPIS-SILO - Built by the GenSpectrum team (cEVO, ETH Zurich)
LAPIS-SILO is a high-performance genomic query engine written in C++. Under the hood, each column value gets a bitmap (one bit per row) indicating which rows contain that value. Queries are answered by bitwise AND/OR-ing the relevant bitmaps together, with Roaring Bitmap compression keeping it compact and cache-friendly.
W-ASAP represents its first application to short read sequencing data - a significant technical milestone that required:
- 88% memory reduction (310GB → 37GB) - proving feasibility despite initial concerns
- Multi-virus support: Path-based routing (
/covid/,/rsva/) for separate databases - Overnight index updates with automatic rollback on failure
Achievements
- 99.77% uptime (December 2025)
- 88% memory reduction enabling cost-effective hosting on Hetzner
- COVID and RSV-A live with daily updates
- From days to minutes: Self-service exploration instead of waiting for expert intermediaries
- Configuration over engineering: Adding new viruses requires only YAML config changes
Live Deployments
COVID Dashboard
genspectrum.org/swiss-wastewater/covid
RSV-A Dashboard
genspectrum.org/swiss-wastewater/rsv-a
LAPIS API
- COVID:
lapis.wasap.genspectrum.org/covid/ - RSV-A:
lapis.wasap.genspectrum.org/rsva/
Resources
ETH Zürich D-BSSE Feature
 Watch on YouTube - Representing Switzerland's national viral wastewater surveillance
Watch on YouTube - Representing Switzerland's national viral wastewater surveillance
Platform Walkthrough
youtu.be/kCUd-o1FbXg - Overview of the platform and analysis capabilities
Workshop Materials
github.com/gordonkoehn/wasap-workshop - Jupyter notebooks for hands-on exploration
API Documentation
- LAPIS COVID: lapis.wasap.genspectrum.org/covid/swagger-ui
- Loculus: db.wasap.genspectrum.org/api-documentation
Team
This work was a collaboration between two groups at ETH Zurich:
CBG (Computational Biology Group, D-BSSE):
- Gordon J. Köhn (me) - Software Engineering, sr2silo, WisePulse, V-Pipe Scout
- Ivan Topolsky - V-Pipe lead, overall technical ownership
cEVO (Computational Evolution, D-BSSE):
- Chaoran Chen - GenSpectrum infrastructure
- Felix Hennig - GenSpectrum dashboards, Loculus deployments, monitoring
- Alexander Taepper - LAPIS-SILO optimization, data architecture
What's Next
The platform continues under CBG and cEVO. The roadmap includes:
- RSV-B pipeline and dashboard (configs ready)
- Influenza integration (pending metadata resolution)
- Expanded COVID lookback window (currently 4 months)
- Usage analytics for stakeholder reporting
The infrastructure I built at ETH Zurich transformed wastewater surveillance from an expert-only cluster workflow into a self-service browser tool. It's live, it's stable, and it's serving Switzerland's public health community.